Running Preprocessing Pipelines
How it works
Whole-slide images are typically too large to load in memory, and computational requirements scale poorly in image size.
PathML therefore runs preprocessing on smaller regions of the image which can be held in RAM,
and then aggregates the results at the end.
Preprocessing pipelines are defined in Pipeline objects.
When SlideData.run()
is called, Tile objects are lazily extracted from the slide by the
SlideData.generate_tiles() method and passed to the
Pipeline.apply() method, which modifies the tiles in place.
Finally, all processed tiles are aggregated into a single h5py.Dataset array and a PyTorch Dataset is generated.
Each tile is processed independently, and this data-parallel design makes it easy to utilize computational resources and scale up to large datasets of gigapixel-scale whole-slide images.
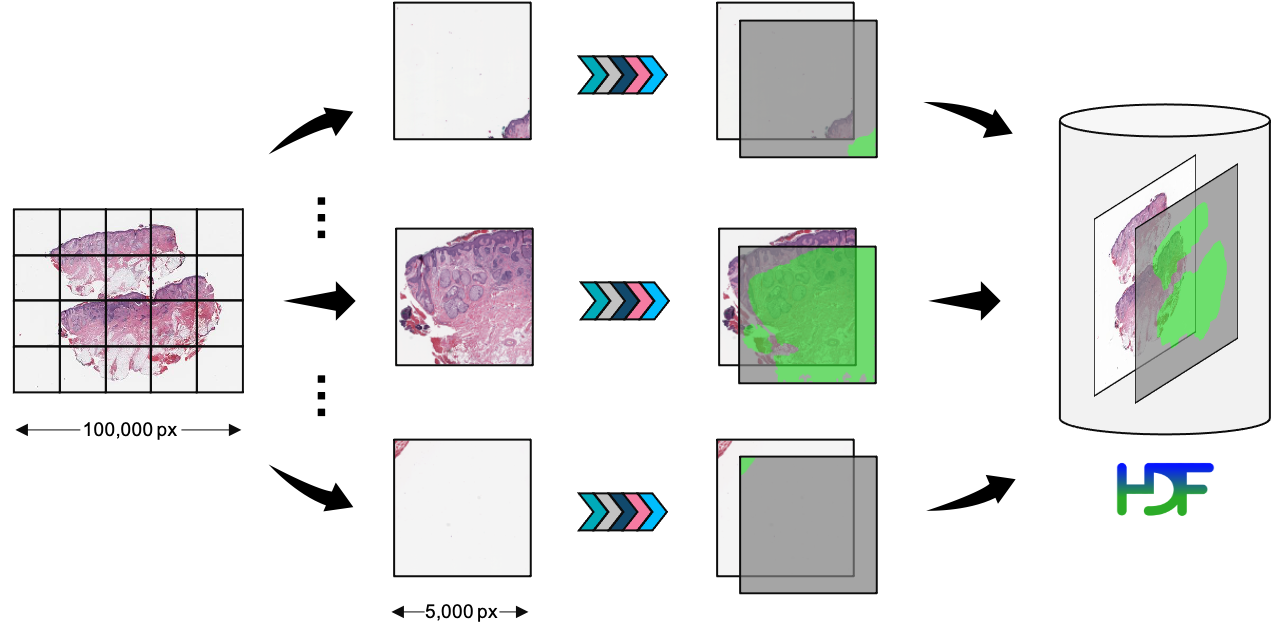
Schematic diagram of running a preprocessing pipeline. Tiles are extracted from the WSI and processed in parallel, before finally being aggregated into a HDF5 file on disk.
Preprocessing a single WSI
Get started by loading a WSI from disk and running a preprocessing pipeline:
from pathml.core import HESlide
from pathml.preprocessing import Pipeline, BoxBlur, TissueDetectionHE
wsi = HESlide("../data/CMU-1.svs", name = "example")
pipeline = Pipeline([
BoxBlur(kernel_size=15),
TissueDetectionHE(mask_name = "tissue", min_region_size=500,
threshold=30, outer_contours_only=True)
])
wsi.run(pipeline)
Preprocessing a dataset of WSI
Pipelines can also be run on entire datasets, with no change to the code:
Here we create a mock SlideDataset and run the same pipeline as above:
from pathml.core import HESlide, SlideDataset
from pathml.preprocessing import Pipeline, BoxBlur, TissueDetectionHE
# create demo dataset
n = 10
slide_list = [HESlide("../data/CMU-1.svs", name = "example") for _ in range(10)]
slide_dataset = SlideDataset(slide_list)
pipeline = Pipeline([
BoxBlur(kernel_size=15),
TissueDetectionHE(mask_name = "tissue", min_region_size=500,
threshold=30, outer_contours_only=True)
])
slide_dataset.run(pipeline)
Distributed processing
When running a pipeline, PathML will use multiprocessing by default to distribute the workload to
all available cores. This allows users to efficiently process large datasets by scaling up computational resources
(local cluster, cloud machines, etc.) without needing to make any changes to the code. It also makes it feasible to run
preprocessing pipelines on less powerful machines, e.g. laptops for quick prototyping.
We use dask.distributed as the backend for multiprocessing. Jobs are submitted to a Client, which takes care of
sending them to available resources and collecting the results. By default, PathML creates a local cluster.
Several libraries exist for creating Clients on different systems, e.g.:
dask-kubernetes for kubernetes
dask-jobqueue for common job queuing systems including PBS, Slurm, MOAB, SGE, LSF, and HTCondor typically found in high performance supercomputers, academic research institutions, etc.
dask-yarn for Hadoop YARN clusters
To take full advantage of available computational resources, users must initialize the appropriate Client
object for their system and pass it as an argument to the SlideData.run() or SlideDataset.run().
Please refer to the Dask documentation linked above for complete information on creating the Client
object to suit your needs.